Research Projects
The overexpression of an intrinsically disordered protein is sufficient to cause AML.
Meningioma-1 (MN1) overexpression in AML is associated with poor prognosis, and forced expression of MN1 induces leukemia in mice. We sought to determine how MN1 causes AML. We found that overexpression of MN1 can be induced by translocations that result in hijacking of a downstream enhancer. Structure predictions revealed that the entire MN1 coding frame is disordered. We identified the myeloid progenitor specific BAF complex as the key interaction partner of MN1. MN1 over-stabilizes BAF on enhancer chromatin, a function directly linked to the presence of a long polyQ-stretch within MN1. BAF over-stabilization at binding sites of transcription factors regulating a hematopoietic stem/progenitor program prevents the developmentally appropriate decommissioning of these enhancers and results in impaired myeloid differentiation and leukemia. Beyond AML, our data detail how the overexpression of a polyQ protein, in the absence of any coding sequence mutation, can be sufficient to cause malignant transformation.
Read our most recent publication about this topic in Molecular Cell!
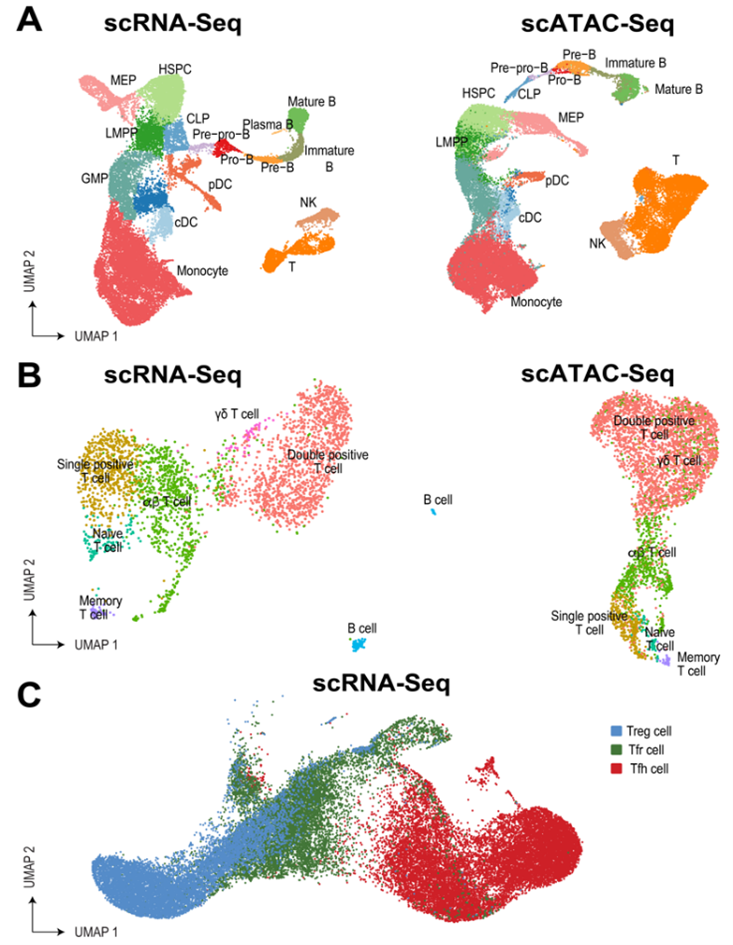
CHIDA - Childhood Hematopoiesis and Immune Development Atlas (Human Cell Atlas - HCA)
The first years of life are critical for establishing protective immunity without inducing allergy, atopy, autoimmune disease, or malignant transformation. The Childhood Hematopoiesis and Immune Development Atlas (CHIDA) will map this complex process at a single-cell resolution. Specific attention will be paid to early development, where some of the most critical changes occur. In addition, the team will aim to over-represent donors of historically excluded ancestries. This is particularly important as underrepresented populations bear a disproportionate burden of disease and face worse outcomes.
Open source computational tools will be used to create dynamic, multi-omic maps of the developing hematopoietic and immune system at both spatial and temporal resolution. In addition, metabolic labeling and signal transduction network inference will allow constructing networks of cellular crosstalk within and across organs as well as their dynamic regulation in the developing child.
Learn more about the remarkable team leading this initiative here.
Single-cell multiomic analysis of pediatric cancers reveals novel prognostic subgroups and mechanisms of disease/resistance.
The incidence and molecular make-up of pediatric cancers is strongly dependent on age. Furthermore, age is a critical predictor of outcome, in some cases even within defined cytogenetic groups. We are contributing to a series of studies that use transcriptomics and epigenomics to better understand pediatric cancers. Read more about this topic here:
Novel prognostic subgoups in T-ALL. (Nature 2024)
Discovery of metabolic alterations in Beckwith-Wiedeman Syndrome and link to cancer (BioRx 2024).
The impact of age on the biology of infant ALL (Blood 2022)
Single cell analysis of rare cells that survive chemotherapy in pediatric cancers (Human Tumor Atlas - NCI Cancer Moonshot)
As part of the Human Tumor Atlas Network (HTAN), the Tan lab has partnered with the Bernt lab to peform a multi-omic single cell analysis of ALL blasts before and after induction chemotherapy, focusing on patients with induction failure or high minimal residual disease (MRD) in an attempt to understand the mechanism that drive resistance and poor outcome.
Tumor transcriptomes/epigenomes uploaded into HTAN
Novel mechanism of chemotherapy resistance in high risk neuroblastoma (Cancer Discovery 2024)
Examining Properties of and Therapies for Malignancies with high HOXA expression.
Overexpression of the HOXA cluster is observed in AML, B-ALL and T-ALL. We are exploring the efficacy and mechanisms of resistance of targeted inhibitors such as Menin inhibitors, ENL inhibitors and DOT1L inhibitors in these diseases. Past contributions and ongoing projects include:
KMT2A-fusion driven AML (10% of AML) requires the histone methyltransferase DOT1L. (Cancer Cell)
KMT2A drive HOXA cluster overexpression in AML without KMT2A fusions. This recognition opened the door for the use of targeted agents designed to interfere with KMT2A-fusions to the much larger group of leukemias with high HOXA cluster expression (60% of AML). (JCI)
ENL inhibitors have in vivo activity in high HOXA expressing AML (KMT2A fusion, NPM1c) (Cancer Discovery)
Menin Inhibition caused due to prior lines of non-menin inhibitory therapies (Experimental Hematology)
The Bernt lab is researching molecular mechanisms in leukemia, utilizing a wide range of models, including:
- Genetically engineered murine models for common leukemogenic drivers (gain of function and loss of function): Inv(16), FLT3-ITD, mIDH2, CDKN2A-, EZH2-, EED, GATA2-
- Retroviral murine models: MLL-AF9, MLL-AF6, MLL-ENL, AML-ETO, MN1, oncogenic NRAS, mIDH1, mIDH2
- Cell lines
- Patient samples and patient derived xenografts.
We study epigenetic and transcriptional regulation in leukemia with the goal of uncovering molecular mechanisms and developing new therapies and combinations.
Visit pubmed for our full list of publications!
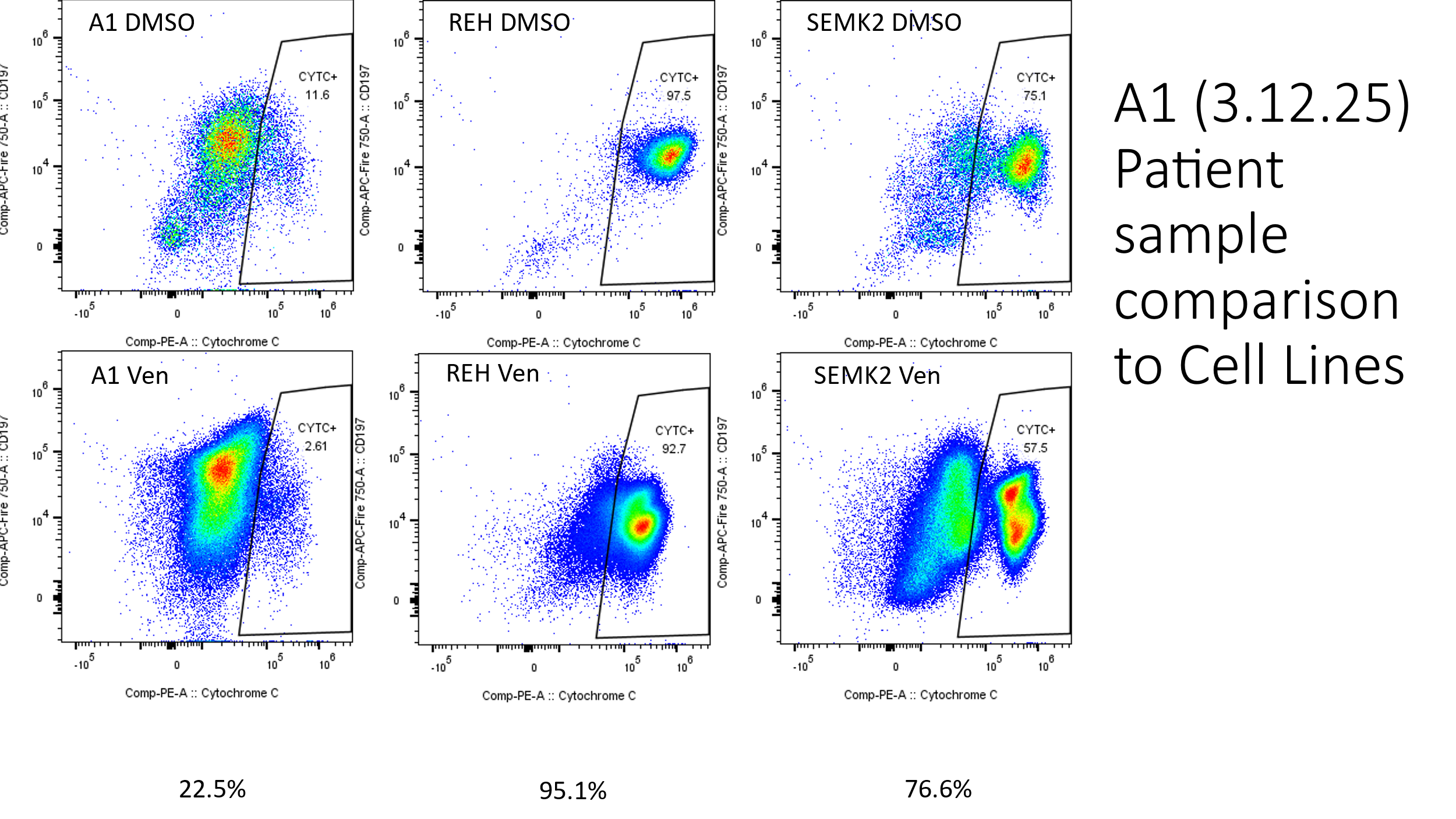
Characterizing Impact of BCL-2 Inhibitors in infant ALL prior to immunotherapy
To reduce relapse and refractory disease in infant ALL (iALL) the Children’s Oncology Group Clinical Trial ALL2321 is comparing the addition of Venetoclax, a BCL-2 inhibitor, to the line of therapy prior to immunotherapy. The Bernt Lab worked with collaborators at the University of Chicago to develop a spectral flow immunophenotyping panel in addition to employing BH3 profiling of the immune compartment at three therapeutic timepoints to evaluate apoptotic dependencies of the iALL and different immune cell types before and after treatment and track those with immunotherapy response and outcome. In addition to these models the Bernt Lab is working with collaborators to perform Whole Genome Sequencing, RNA-Sequencing, and other analysis on these samples to fully characterize these samples.